Release date: 2014-10-20
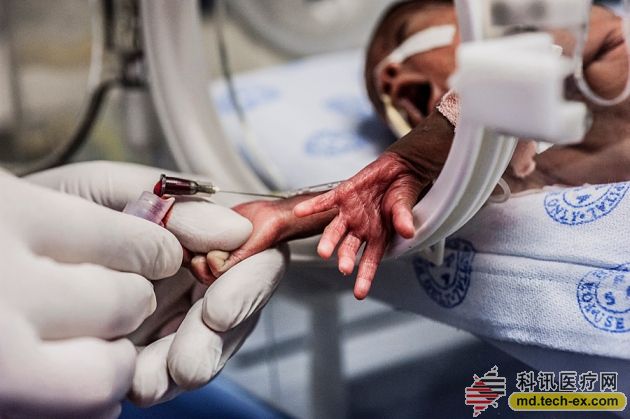
The genome of the diseased newborn will be sequenced within 24 hours, providing clinicians with a quick diagnostic basis.
When only two months old, the boy was already on the line. His entire short life was spent in the neonatal intensive care unit at the Children's Charity Hospital in Kansas City, Missouri. The physicians racked their brains to find the cause of their abnormal condition. In April 2013, the baby's liver began to fail, and the medical staff told his parents that the prospects were not optimistic.
Subsequently, geneticist Stephen Kingsmore from the Children's Charity Hospital and his team took over the patient. In three days, they sequenced the baby's and their parents' genomes and confirmed a rare mutation common to both his and his parents. This mutation has been shown to be associated with a disease caused by an overreactive immune system that can damage the liver and spleen. With such a diagnosis, a baby boy's physician has the right medicine to reduce his immune response. Today, the patient has returned home and is in good health.
The Kingsmore team has sequenced the genomes of 44 sick babies by using a process that provides diagnostics within 24 hours. The baby boy is just one of them. In 28 cases, the researchers were able to diagnose the condition of these babies. At the same time, in about half of the cases, they suggested making adjustments in treatment methods. Kingsmore recently reported the results of the study at the Common Disease Genomics Conference in Maryland, USA. On October 6, his team launched a larger project to sequence the genomes of hundreds of babies. This will be the first of four neonatal sequencing studies, and each of the previous studies received multi-million dollar funding from the National Institutes of Health (NIH) in September 2013. These studies will address the feasibility and ethical issues of the above process as it will soon become a common standard for newborns with unexplained conditions.
Over the next five years, the Kingsmore team will sequence the genomes of 500 sick babies in the neonatal intensive care unit at the Children's Charity Hospital, and will use the clinical efficacy of another 500 identical infants diagnosed with traditional genetic and metabolic tests. Comparison. The researchers will also assess whether rapid sequencing can prevent these babies from unnecessary testing and useless treatment, and whether it can help parents make decisions about whether to continue treatment when their child is diagnosed with an incurable disease. Kingsmore said that even if some babies die, genome sequencing and diagnosis can provide more information about the genetic diseases they carry.
Kingsmore refers to this rapid sequencing technique as a "factory" approach in which four or five experts each operate as quickly as possible from one stroke to the final diagnostic process. The research team collects DNA from parents and children to more quickly identify mutations in the child's genome, then sequence the DNA and use custom software to "target" specific parts of the genome based on patient symptoms. After making a gene-based diagnosis and submitting relevant information to the physician's physician, the team will store the sequencing data in a secure database in an anonymous manner for future research use.
Misha Angrist, a genomics policy expert at Duke University in the United States, said that although the 24-hour genome sequencing process is impressive, it is unclear whether gene sequencing for newborns can quickly become a common practice. A number of issues remain to be resolved, such as who pays for sequencing, who can access relevant data, and how clinicians can grasp the scale when extracting genomic information unrelated to current diseases.
So far, only the Kansas City research team has started the trial, thanks to the US Food and Drug Administration (FDA) exemption – agreeing to sequence the sickly babies. Usually a test must be proven by experiment before being used to diagnose a patient. "I think everyone is very keen to see if this will be the beginning of a new FDA sequencing method and whether it will appear in similar trials in the future," Kingsmore said.
Other NIH-funded teams are awaiting approval from the FDA or the internal ethics review board. In Boston, a team led by Boston Children's Hospital physician Alan Beggs and Brigham and Women's Hospital physician Robert Green is planning a study of 240 healthy babies and 240 neonatal intensive care units. The team will randomly sequence half of the genome-encoded protein portion of each group of infants, the exome, to determine whether these data alone can improve the health of the baby.
A third team led by geneticists Cynthia Powell and Jonathan Berg of the University of North Carolina at Chapel Hill plans to sequence 400 infants with known genetic diseases such as cystic fibrosis to see if additional disease-related episodes can be collected. information. A team led by medical geneticist Robert Nussbaum of the University of California will sequence the exome of 1400 blood drops collected at the time of birth to determine whether this information can help diagnose.
Each team includes ethicists who will work to resolve issues such as ensuring that information that is not related to the diagnosis is not disclosed. "People are very sensitive to the power of genomics information, and the truth is also true." Green said that these concerns would be magnified several times when it comes to babies.
Source: China Science and Technology News
Disposable Tracheostomy Tube Kit
Tracheostomy Tube Kit,tracheostomy tube,pediatric tracheostomy tube 4.5,shiley tracheostomy tube
Anesthesia Medical Co., Ltd. , https://www.medicaldiverse.com